

RAPID Platform
A universe of therapeutic
possibility, within reach.
Jnana has built a next-generation chemoproteomics platform to develop small molecule medicines for any target class with unprecedented speed and efficiency.

Early advances in chemoproteomics have shed new light on previously inaccessible therapeutic targets, illuminating binding sites undetected by conventional approaches. Yet still, small molecule drug discovery has remained slow and inefficient, and many disease targets have remained elusive. Until now.
Powered by next-generation chemoproteomics and a world-class team, Jnana has reimagined drug discovery at every stage. We start with well-validated targets – those that human biology and genetics have revealed are significant drivers of disease. Then we deploy our
RAPID chemoproteomics platform to drug these high value targets with improved efficiency, flexibility and speed.
With RAPID, we can address a range of challenging target classes, including SLC transporters, transcription factors, signaling scaffold proteins, phosphatases, GPCRs and helicases. Our binding-based screening approach is inherently flexible – enabling the identification of small molecules that elicit diverse pharmacologies, such as allosteric inhibitors, modulators of protein localization or molecular glues.
The RAPID Platform
SELECT THE TARGET
FIND THE POCKETS
SCREEN FOR BINDERS
RAPID is performed in living cells, providing functionally relevant data in native biological contexts while allowing the interrogation of protein targets that might otherwise be challenging to isolate and purify for screening.
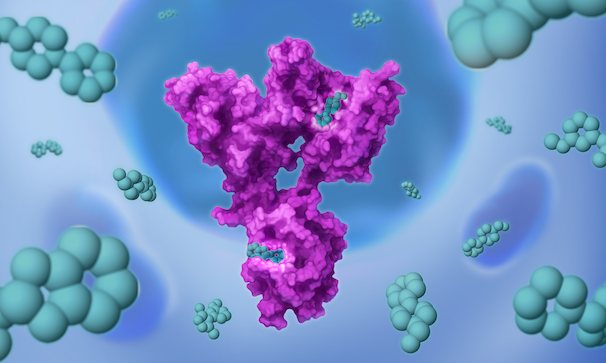
Once we’ve set our sights on a biologically validated target, we unleash Jnana’s proprietary Reactive Affinity Probe (RAP) library – a collection of chemical fragments that can bind and covalently label any druggable pocket on a target. Designed to cover the diverse spectrum of chemical space, our growing library of RAPs is continually optimized based on success criteria – maximizing our ability to uncover novel binding sites on hard-to-drug targets.
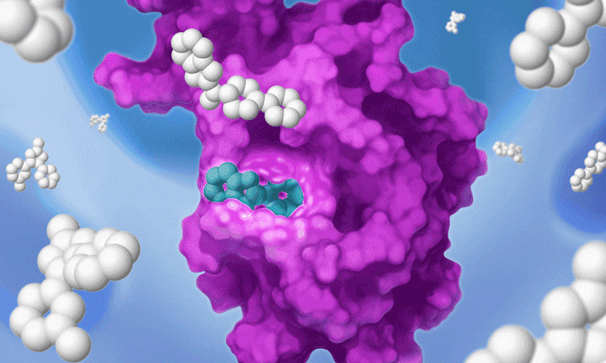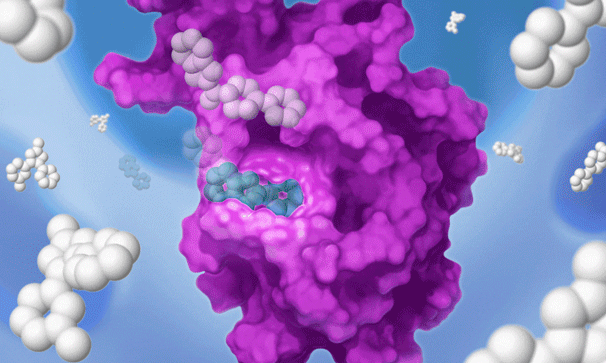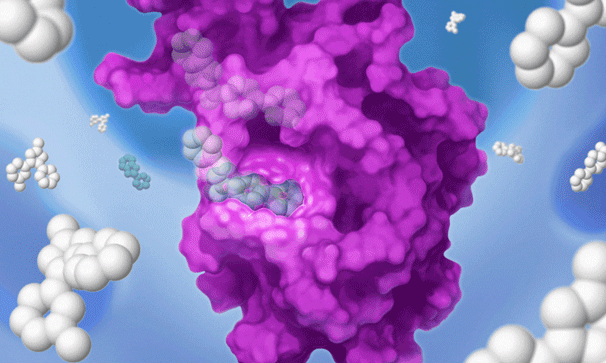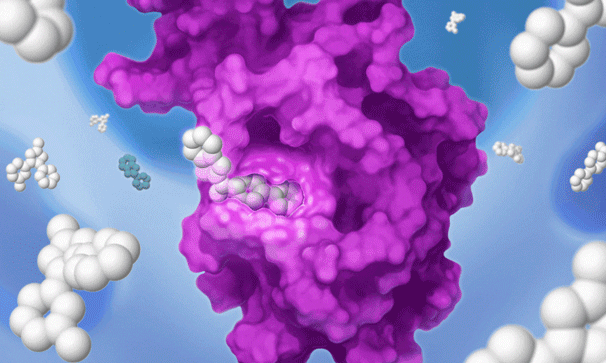
Once we identify a RAP that binds to a pocket on the target of interest, we can use our proprietary detection technology to run a high-throughput screen of a large library of drug-like compounds to quickly identify small molecules that bind to the target in cells and prevent the RAP from binding and labeling the target. This efficient high-throughput screening approach circumvents the need for iterative structure-based fragment optimization, significantly expediting the time it takes to arrive at drug-like leads.
